There are several ways to utilize the AcaFinder website:
Welcome to AcaFinder! The Home page features an introduction to AcaFinder that discloses all incorporated bioinformatic tools, and a detailed outline of the AcaFinder workflow in a figure.
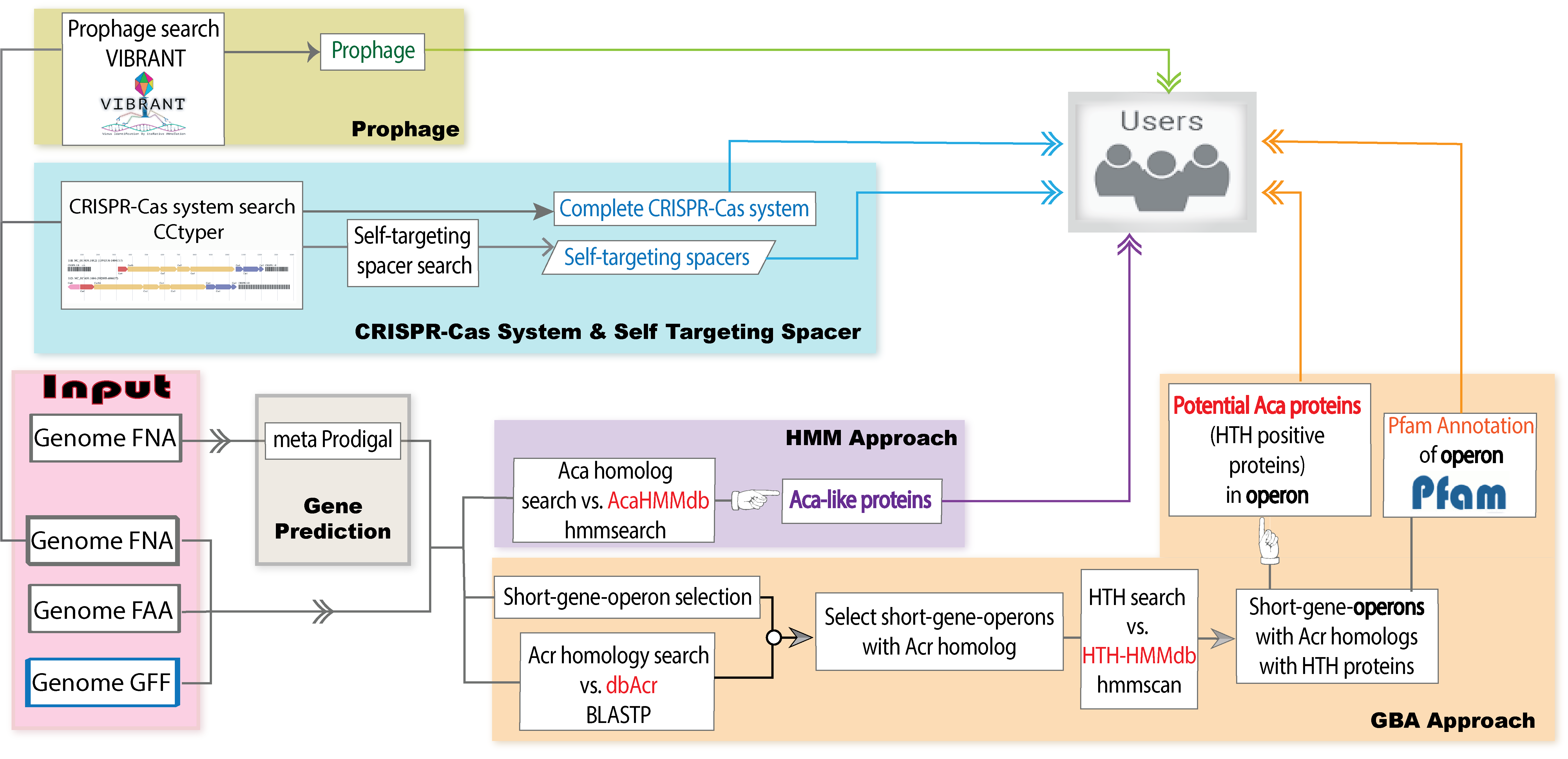
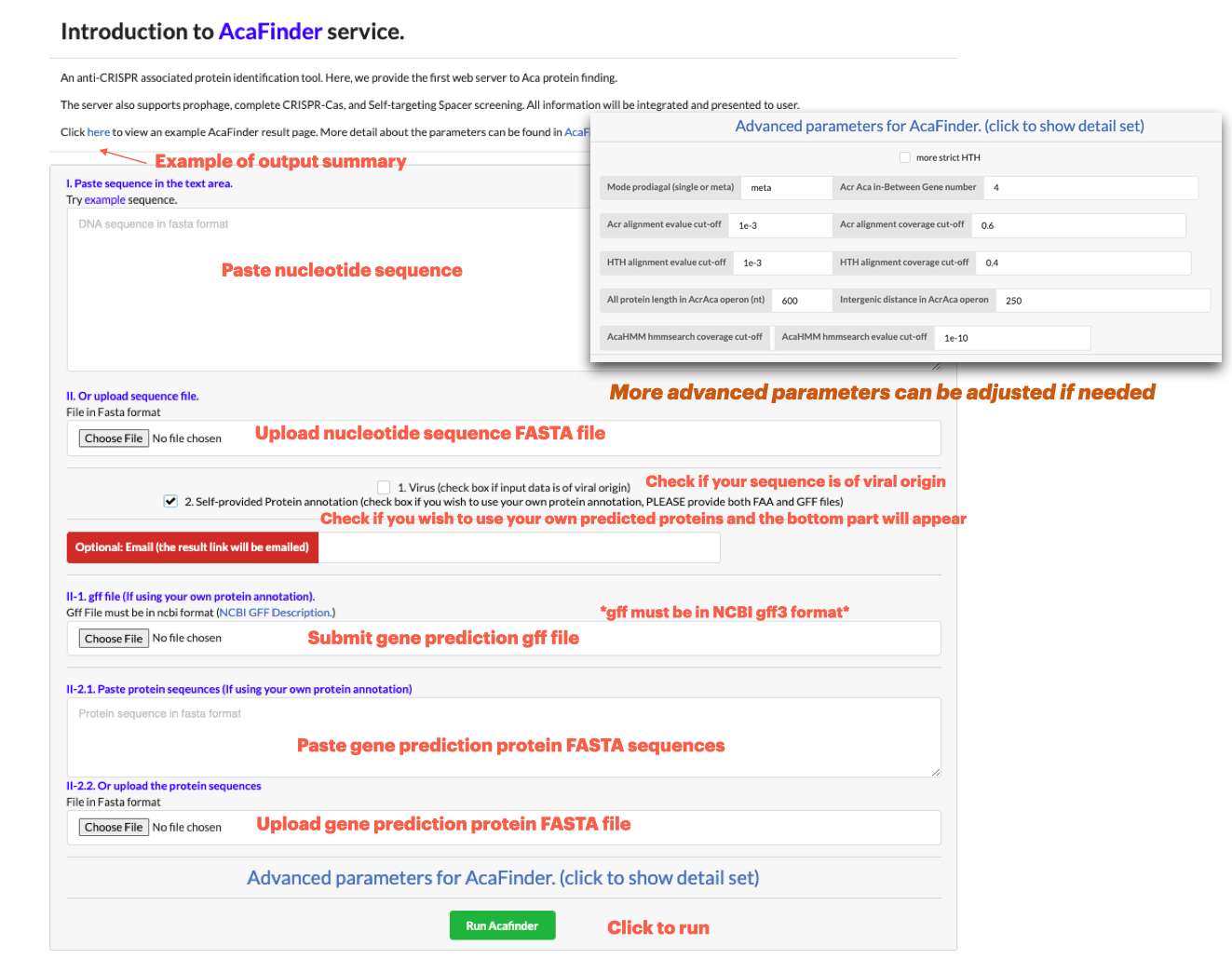
To input a genomic sequence for Aca screening, go to Run AcaFinder on the navigation bar. Sequence data can be submitted in two ways:
**Data of ONE genome may be submitted at a time**
**Higher genomic assembly quality preferred**
Results are then displayed, where users can view where the Aca protein/operons are, and their associatin with prophages as well as CRISPR-Cas systems (if any)
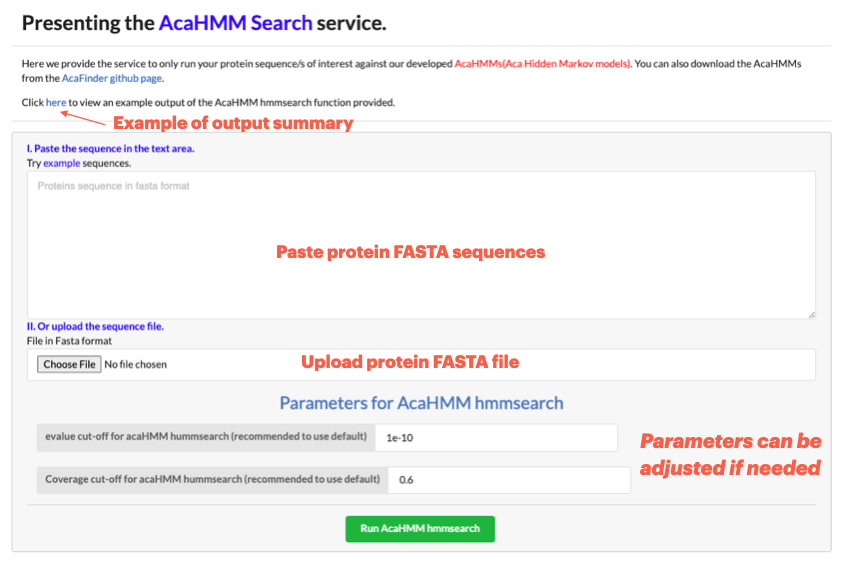
Go to Run HMMsearch to only run your protein sequence/s of interest against our developed AcaHMMs(Aca Hidden Markov models)
The output of AcaFinder consists of the following:

Copyright 2022 © YIN LAB, UNL. All rights reserved. Designed by Bowen Yang and Jinfang Zheng. Maintained by Yanbin Yin.