

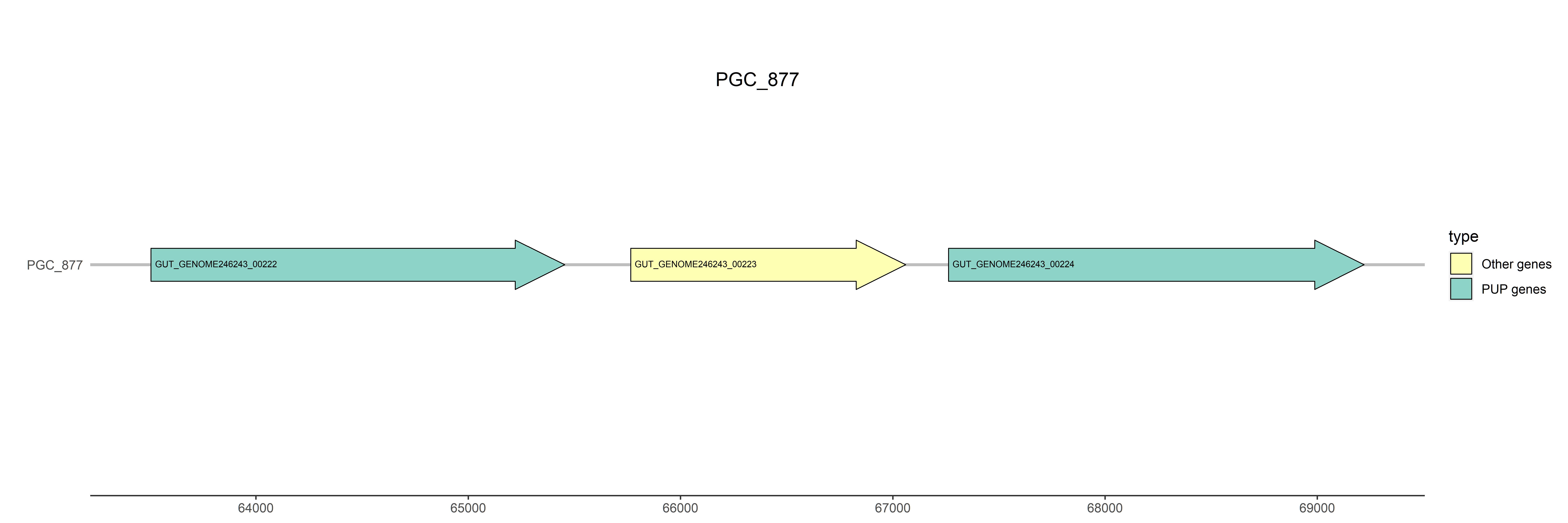
Gene: NA
Representative genome: GUT_GENOME246243
Gene type: PUP genes
Position: 63505..65454
Protein length: 650
Product: NADH oxidase
EC: NA
Pfam: FMN_red, FMN_bind, FAD_binding_2
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Clostridium_Q; Clostridium_Q symbiosum
Geography: Fiji, Oceania
Gene: NA
Representative genome: GUT_GENOME246243
Gene type: other genes
Position: 65765..67060
Protein length: 432
Product: HTH-type transcriptional activator RhaS
EC: NA
Pfam: FMN_red, FMN_bind, FAD_binding_2
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Clostridium_Q; Clostridium_Q symbiosum
Geography: Fiji, Oceania
Gene: NA
Representative genome: GUT_GENOME246243
Gene type: PUP genes
Position: 67262..69220
Protein length: 653
Product: NADH oxidase
EC: NA
Pfam: Pyr_redox_2, Oxidored_FMN, FGGY_N, FGGY_C, cNMP_binding, HTH_Crp_2
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Clostridium_Q; Clostridium_Q symbiosum
Geography: Fiji, Oceania